Table of Contents
The development of methods of DNA sequencing has contributed to the study of organisms’ genomic sequences. Maxam and Gilbert found the binding site for lactose-repressors by copying and sequencing 24 bases. Sanger and Maxam-Gilbert procedures were more widespread previously and are still used to determine the order of the nucleotide bases adenine, guanine, cytosine, and thymine in a DNA molecule. Early attempts to sequence DNA were laborious.
However, technological advancements have massively eased the process as of now. Although, with today’s technological improvements, most of the digital instruments are largely based on them. DNA sequencing techniques are divided into three categories.
First Generation Sequencing
Sanger and Maxam-Gilbert sequencing have been designated as First Generation Sequencing Technologies. Many methods for similar sequencing have been devised; nevertheless, only small DNA fragments (less than one kilobase) can be sequenced.
Sanger sequencing
Sanger sequencing is sometimes known as chain termination sequencing, dideoxynucleotide sequencing, or synthesis sequencing. It involves utilizing a single strand of double-stranded DNA as a template for sequencing. This sequencing is carried out using chemical nucleotides known as dideoxy-nucleotides. Once incorporated into the DNA chain, these nucleotides act as dNTPs for nucleotide elongation, blocking further extension, and the elongation is completed. Following that, DNA fragments of varied sizes that have been treated with dNTP are collected.
Using a gel block, the fragments are separated by their size such that an imaging device may be observed with the succeeding bands corresponding to DNA fragments. Even now, Sanger sequencing has been widely utilized for single or low-throughput DNA sequencing for many decades. However, developing a processing rate that does not require amplification of the large DNA is still challenging.

Maxam-Gilbert Sequencing
Maxam-Gilbert depends on chemical cleaving of nucleotides, which works best with small polymers of nucleotides. Chemical treatment introduces breaks in each reaction at a restricted fraction of one or two of the four nucleotide bases. This process produces a collection of identifiable particles that can be separated by electrophoresis based on their size. Maxam and Gilbert sequenced either double or single-stranded DNA strands that were labeled at one end of a 32P chain.
Chemical treatment caused breakage at a low fraction of one or two of the four nucleotide bases in each of four reactions. As a result, from the radiolabeled end to the first cut site in each molecule, a series of identifiable fragments is formed. The fragments are then quantified using gel-electrophoresis and authoradiography, yielding a representation of bands relating to the radiolabeled fragments from which the sequence is inferred.

Second Generation Sequencing
For three decades, first-generation sequencing, particularly Sanger sequencing, dominated; nonetheless, cost and time were important issues. The advent of a new generation of sequencers in 2005 broke the limits of first-generation sequencers. The following are the fundamental elements of second-generation sequencing technology:
- The generation of many millions of short reads in parallel
- The speedup of sequencing the process compared to the first generation
- The low cost of sequencing and
- The sequencing output is directly detected without the need for electrophoresis
- Shotgun sequencing of randomLy fragmented genomic DNA without the need of cloning via a foreign host cell
- Library amplification is performed on a solid surface or beads within miniature emulsion droplets
Short read sequencing methods separated into two side approaches: ligation sequencing, and synthesis sequencing. Because a high-throughput sequencing is readily feasible, sequencing of the second generation is also known as Next-Generation Sequencing ( NGS). Different platforms based next-generation sequencing are 454, ABI SOLiD, Illumina, and Semiconductor sequencing (Ion Torrent).
Roche/454 Sequencing
In 2005, a company named 454 Life Sciences Corporation took the first steps in the NGS movement. The name 454 derived from this business which was later purchased by Roche in 2007. The Concept behind 454 is focused on pyrosequencing, which was a Pyrosequencing Ab approved technology. This method depended on the inorganic pyrophosphate (PPi) being produced during PCR when a complementary base was inserted. The sequencing reaction happens very quickly, in the range of milliseconds, and a charging-couple-device ( CCD) camera may detect the light generated.
The use of a picotiter plate allowed hundreds of thousands of reactions to take place in parallel and was able to generate reads for around 500-600 base pairs long for the millions or so of wells that would be required to contain sufficiently clonally-coated bead. The 454 technological breakthrough miniaturized the sequencing reaction, enabling simultaneous sequencing reactions to occur in a limited space utilizing a smaller number of reagents. The newer version of the 454 instruments will produce reads up to 1000 bp in lengths, generating millions of reads per cycle.
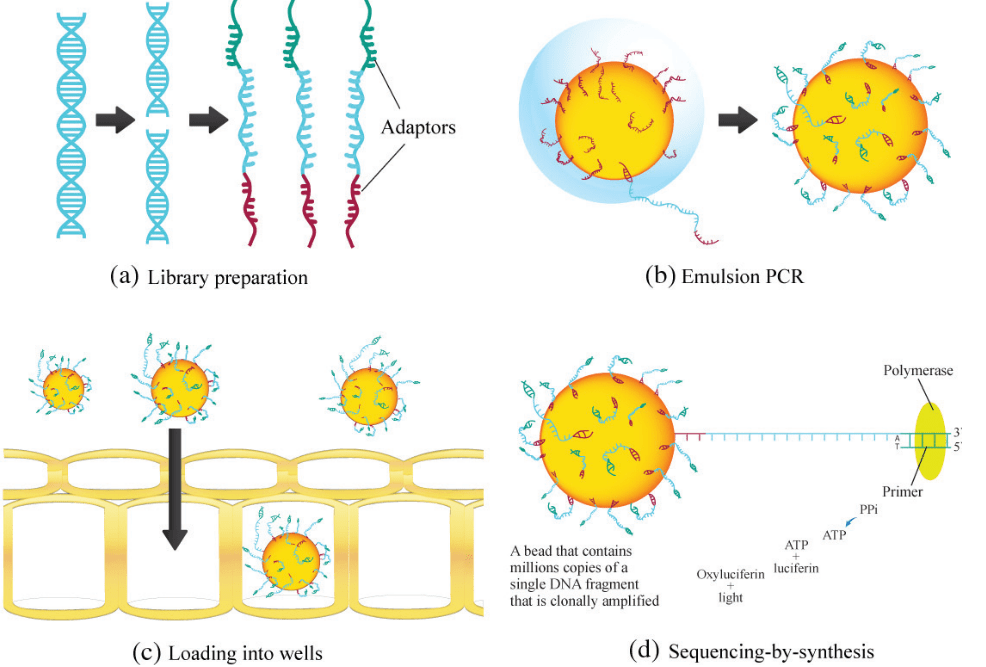
Description of Figure:
- 454 Pyrosequencing, procedure initiated with a single-stranded library with adaptors ligated on both ends.
- The adaptor binds to the beads, which is further subjected to emulsion PCR, which generates millions of copies of a single DNA fragment on each bead.
- The beads are placed into PicoTiter Plate for sequencing by detection of base incorporation during PCR.
- When a nucleotide base is incorporated, inorganic pyrophosphate (PPi) is generated, which is converted to ATP by sulfurylase and luciferase uses the ATP to oxyluciferin and light.
The 454 ‘s crucial drawback is the difficulties of measuring the real amount of homopolymer tract bases (e.g., TTTTT). It lacked blocking mechanism to avoid the inclusion of several similar bases during elongation of DNA. It was difficult to determine how many bases there are until the homopolymer is longer than eight bases, only more accurate light signals flow meaning. Signals of too large or too low amplitude led to the amount of nucleotide bases being underestimated or overestimated, which triggered irregular nucleotide recognition.
ABI SOLiD
In 2006, ABI acquired the takeover of Agencourt Personal Genomics, allowing the deployment of revolutionary NGS technology known as Supported Oligo Ligation Detection (SOLiD). The Strong sequencing system is problematic due to the di-base encoding mechanism. The sample preparation measurements prior to probe ligation are quite similar to 454 programs. The adaptors are ligated to previously fragmented DNA and enriched on beads using emulsion PCR. The beads are then covalently attached to a glass slide, with which bases are ligated and detected. A universal sequence-primer is employed to bind to the established adaptor chain.

Description of figure:
- Each ligation cycle starts with the 8-mer probe binding to the template and then ligated for its detection. Then, cleavage occurs to remove three nucleotides and a tagged dye.
- Structure of the 8-mer probe.
- An illustration of the sequence determination process during each ligation cycle of the primer rounds. Position 0 is a part of the adaptor sequence, and the template sequence is only revealed from position one onwards.
The ligation and cleavage cycle may be repeated numerous times to ensure the required sequence duration. While the SOLiD framework is unique in that it can preserve the sequence of oligo color calls to be utilized for mutation calls, this has caused issues for bioinformatics research because most approaches rely on DNA calls rather than color space models.
Illumina
Solexa initially released the first sequencer, Genome Analyzer, and was later purchased by Illumina in 2007. Some of Illumina ‘s crucial strengths is its capacity to produce large DNA sequence data output at reduced expense, while generating only short sequences. In addition to the beneficial low-cost high-performance sequencing, Illumina does higher than the 454 homopolymer sequencing method, because it employs reversible terminator sequencing chemistry.

Description of figure:
- Genomic DNA is sheared, size selected, and then attached with adaptors at both ends.
- DNA library, placed on the flowcell to allow for complementary binding at one end of the adaptor to probes that are coated on the surface. The solid-phase bridge amplification generated clusters of single DNA fragments. Reverse strands cleaved and washed away.
- Sequencing initiated with primer addition to the remaining forward strand, further addition with the incorporation of one nucleotide at a time.
Illumina devices come with drawbacks too. The sequence end of 3′ appears to be of poorer quality than the end of 5′ which ensures that any sequences from the end of 3′ will be filtered out because it is beyond a certain defined level. Sequence-specific errors for twisted repeats and GGC sequences have also been identified (Low and Tammi, 2017).
Ion Semiconductor Sequencing
Semiconductor sampling, also known as Ion-Torrent sampling, was developed by Life Technologies and later introduced by Thermo Fisher Scientific. It was equivalent to 454 pyrosequencing technology since it did not utilise fluorescent-labeled nucleotides, unlike other second-generation approaches.
The Ion Torrent system, like the 454 and SOLiD methods, incorporates library preparation and emulsion PCR procedures on the beads. The fundamental characteristic is that the sequencing of nucleotides is identified by calculating the H+ ions created during the cycle rather than fluorescence or chemiluminescence. A nanoscale pH-sensor semiconductor made nucleotide identification possible. Each of the four DNA bases is delivered sequentially for DNA elongation; if the base matches the reference, a signal is obtained and recorded correspondingly.
The homopolymer area in the signal should be improved, it is difficult to detect the true number of bases (more than six bp), which causes insertion and deletion errors at a rate of roughly 1%. However, several ways have been tried to solve the constraints of the Ion Torrent technology, including refinement by the bioinformatics system.

Description of the figure:
- The addition of every new nucleotide leads to the release of H+.
- A silicon pH sensor detects the release of H+.
- There exist several millions of pH sensors.
- pH sensors are arranged within a sequencing chip.
DNB sequencing
DNB sequencing technology is changing genomics for the better. It helps us analyze DNA faster, more accurately, and at a lower cost. This cool method uses something called DNA nanoballs (DNBs) to do the job better than other techniques. With DNB sequencing, we put individual DNA pieces inside tiny nanoballs, and this lets us read lots of DNA really quickly. It’s like a superpower for genetics!
Here’s how it works: We put DNA inside nanoballs and then check them out. Each nanoball has just one piece of DNA, but we can look at lots of nanoballs all at once. This means we can study many DNA pieces at the same time, which is fantastic for things like studying different groups of people or researching cancer. DNB sequencing is super precise and sensitive, too. It hardly makes any mistakes and can read long pieces of DNA. It’s a must-have tool for scientists all over the world, making genetics research exciting and accessible to everyone.
DNB sequencing is a game-changer in genetics. It can handle DNA inside nanoballs, making it perfect for quick and accurate DNA reading. This incredible technology has tons of uses, from tailoring treatments to studying populations and fighting cancer. DNB sequencing is fast, affordable, and can be used on a large scale. It’s speeding up our understanding of the human genome. As we explore its potential, DNB sequencing is opening doors to better diagnoses, targeted treatments, and improved health. Embracing this technology means genetics will be a big part of the future of healthcare and beyond.

Third-Generation Sequencing
Despite its popularity, second-generation sequencing often requires the PCR amplification stage, which is a time-consuming approach. Furthermore, second-generation sequencers are incapable of dealing with the different genomes of repeated sequences. The third generation of sequencing emerged in response to the problems posed by second-generation sequencers. Third-generation sequencing technologies offer low-cost sequencing and direct sample preparation without the need for PCR amplification, far faster than second-generation sequencing technologies. Third-generation sequencing is described using two major approaches.
- The single-molecule real-time sequencing approach (SMRT)
- The synthetic approach relies on existing short-read technologies.
The most widely used sequencing platforms in third-generation sequencing are MinION and Oxford Nanopore sequencing.
Single-Molecule Real-Time (SMRT) sequencing
Pacific Bioscience made significant advancements that allowed real-time tracking of DNA synthesis. A fluorescent dye is applied to the phosphate line rather than the nucleus of each nucleotide using phospholinks. During DNA elongation, the phosphate chain is broken, causing the dye signal to move forward. The DNA template can accept another nucleotide. Another significant innovation is the use of the Zero-mode-waveguide (ZMW) as a technique for detecting foundation integration. These ZMWs are built into an SMRT instrument.
ZMW are tens of nanometer-diameter wells microfabricated in a metal sheet, which is then put onto a glass substrate. Each ZMW has a DNA polymerase coupled to its bottom as well as the target DNA sequence for sequencing. The DNA polymerase incorporates fluorescent-labeled nucleotides into the DNA fragment during the sequencing step (with different colours). When a nucleotide is introduced, it releases a bright signal that is detected by sensors.

Oxford Nanopore Sequencing
Oxford Nanopore sequencing was created to identify the order of nucleotides in a DNA sequence. Oxford Nanopore Technologies launched the MinION device in 2014, which could generate longer readings, leading to greater resolution structural genomic variations and repetitive sequences.
The DNA fragment is transported through a synthetic protein nanopore in nanopore sequencing. When the DNA fragment is processed through the pore by the activity of a motor protein connected to the pore, it causes a variation in an ionic current produced by the mobile nucleotides filling the pore. The fluctuation of the the ionic current is recorded gradually on a graphical form and then analyzed to determine the sequence. The ‘template read’ is generated by sequencing the direct strand, while the ‘complement read’ is generated by reading the DNA complementary strand attached to the hairpin.
When the “template read” and “complement read” are combined, the resultant consensus sequence is known as a “two-direction read,” or “2D.” Oxford Nanopore sequencing has the advantages of being economical and lightweight. Sequencing information can be visualized on the screen as the sample is placed into the sequencing port without any delays.

Helicos sequencing
The Helicos sequencing technology, which is presently being commercialized by Helicos Biosciences, was the first industrial use of fluorescent single-molecule sequencing. Seqll sequencing currently use the Helicos sequencing technology and HeliScope single-molecule sequencers. DNA is sheared, tailed with polyA, and hybridized to a flow cell surface containing oligo-dT for billions of molecules of sequential sequencing-by-synthesis.
PolyA-tailed DNA molecules hybridize to the oligo-dT50 affixed to the surface of glass flow cells. The addition of fluorescent nucleotides with an ending nucleotide pauses the cyclic cycle until a single nucleotide signal is recognized for each DNA segment, and then the cycle is reproduced before the fragments are completely sequenced. This sequencing approach combines DNA polymerase sequencing with hybridization and synthesis sequencing.
Because there is no ligation or PCR amplification involved in sample preparation, the GC material and size discrepancies reported in other methods are often removed. HeliScope sequencing read lengths range from 25 to over 60 bases, with a maximum of 35 bases. This method successfully sequenced the human genome [PMID: 19668243] to identify disease markers in clinical assessments, as well as sequencing RNA to provide tissue and cell quantitative transcriptomes.
References:
- Heather, James M, and Benjamin Chain. 2016. “The Sequence of Sequencers: The History of Sequencing DNA.” Genomics 107 (1): 1–8. https://doi.org/10.1016/j.ygeno.2015.11.003.
- Kchouk, Mehdi, Jean Francois Gibrat, and Mourad Elloumi. 2017. “Generations of Sequencing Technologies: From First to Next Generation.” Biology and Medicine 09 (03): 1–8. https://doi.org/10.4172/0974-8369.1000395.
- Kulski, Jerzy K Kulski ED1 – Jerzy K. 2016. “Next-Generation Sequencing — An Overview of the History, Tools, and ‘Omic’ Applications.” In , Ch. 1. Rijeka: IntechOpen. https://doi.org/10.5772/61964.
- Low, Lloyd, and Martti Tammi. 2017. Bioinformatics A Practical Handbook of Next Generation Sequencing and Its Applications. Edited by Lloyd Low and Martti Tammi. Singapore: World Scientific Publishing Co. Pte. Ltd. 5.
- Maxam, Allan M, and Walter Gilbert. 1977. “A New Method for Sequencing DNA.” Proceedings of the National Academy of Sciences of the United States of America 74 (2): 560–64. https://doi.org/10.1073/pnas.74.2.560.
- Shendure, Jay, Shankar Balasubramanian, George M Church, Walter Gilbert, Jane Rogers, Jeffery A Schloss, and Robert H Waterston. 2017. “DNA Sequencing at 40: Past, Present and Future.” Nature 550 (7676): 345–53. https://doi.org/10.1038/nature24286.
- Shin, Saeam, Yoonjung Kim, Seoung Chul Oh, Nae Yu, Seung-Tae Lee, Jong Rak Choi, and Kyung-A Lee. 2017. “Oncotarget 34858 Www.Impactjournals.Com/Oncotarget Validation and Optimization of the Ion Torrent S5 XL Sequencer and Oncomine Workflow for BRCA1 and BRCA2 Genetic Testing.” Oncotarget 8 (21): 34858–66. www.impactjournals.com/oncotarget/.
- Kohn AB, Moroz TP, Barnes JP, Netherton M and Moroz LL (2013) Single-cell semiconductor sequencing. Methods in Molecular Biology. 1048(author Manuscript)
